Multiscalar Data Integration
EURenOmics is in the unique position to generate large-scale multi-dimensional molecular profiles along the genotype – phenotype continuum from deeply phenotyped patient cohorts with several rare renal diseases. Defining dependencies between the molecular data sets and the comprehensive clinical profiles will allow (a) redefinition of the nosology of renal disease towards mechanism based ontologies, (b) identification of predictors for outcome and treatment responses and (c) definition of novel regulatory pathways for targeted intervention. To achieve this goal an iterative process engaging the domain experts from WP 2-6 with the bioinformatics expertise in WP7 is essential. The integrative biology team of WP7 has 10 years of experience (EU FP 5: “Chronic progressive kidney disease”) introducing the European renal research community to concept extraction from large scale data sets in a longstanding public-private partnership with the SME Genomatix. The data analysis and mining pipeline, which has emerged from these efforts and will be implemented in EURenOmics is designed with a large degree of flexibility to allow in a WP specific manner multiple entry and exit points for large-scale data sets, integration into biological context and subsequent presentation of dependencies in intuitive graphical manner for iterative data analysis. For further details of specific pipeline elements see WP7.
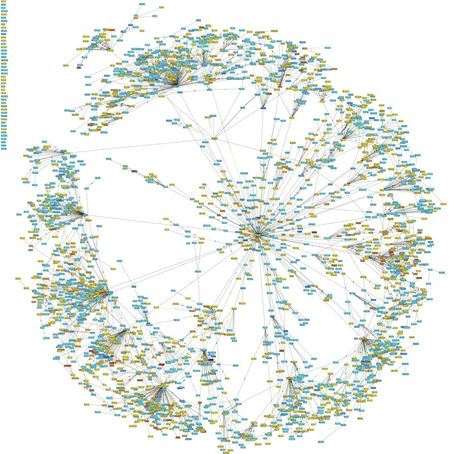
- Figure 1. Example of a disease network established through integrative analysis.